DS4Sci_EvoformerAttention: Code and Tutorial
Model partner: OpenFold team, Columbia University
Introduction
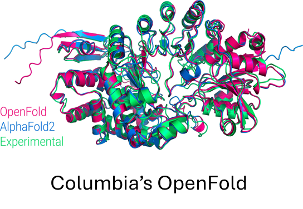
OpenFold is a community reproduction of DeepMind’s AlphaFold2 that makes it possible to train or finetune AlphaFold2 on new datasets. Researchers have used it to retrain AlphaFold2 from scratch to produce new sets of model parameters, studied the early training phase of AlphaFold2 (Figure 1), and developed new protein folding systems.
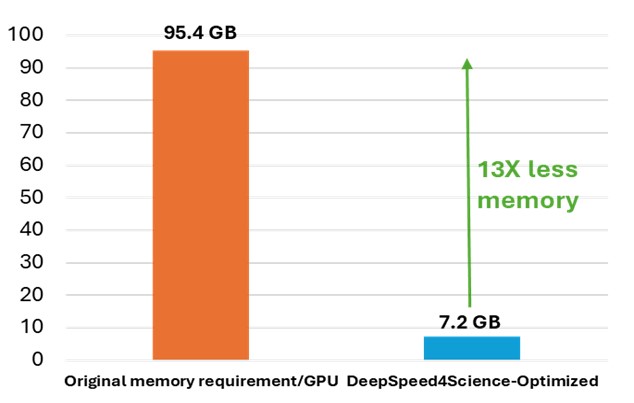
While OpenFold does apply performance and memory optimizations using state-of-the-art system technologies, training AlphaFold2 from scratch is still computationally expensive. The model at the current stage is small in absolute terms, with just 93 million parameters, but it contains several custom attention variants that manifest unusually large activations. During the “finetuning” phase of a standard AlphaFold2 training run, the logit tensor produced in just one of these variants–one designed to attend over the deep protein MSAs fed to the model as input–is in excess of 12GB in half precision alone, dwarfing the peak memory requirements of comparably sized language models. Even with techniques like activation checkpointing and DeepSpeed ZeRO optimizations, this memory explosion problem heavily constrains the sequence lengths and MSA depths on which the model can be trained. Furthermore, approximation strategies can significantly affect the model accuracy and convergence, while still resulting in memory explosion, shown as the left bar (orange) in Figure 2.
To address this common system challenge in structural biology research (e.g., protein structure prediction and equilibrium distribution prediction), DeepSpeed4Science is addressing this memory inefficiency problem by designing customized exact attention kernels for the attention variants (i.e., EvoformerAttention), which widely appear in this category of science models. Specifically, a set of highly memory-efficient DS4Sci_EvoformerAttention kernels enabled by sophisticated fusion/tiling strategies and on-the-fly memory reduction methods, are created for the broader community as high-quality machine learning primitives. Incorporated into OpenFold, they provide a substantial speedup during training and dramatically reduce the model’s peak memory requirement for training and inference. This allows OpenFold to be experimented with bigger and more complex models, and longer sequences, and trained on a wider spectrum of hardware.
Methodology
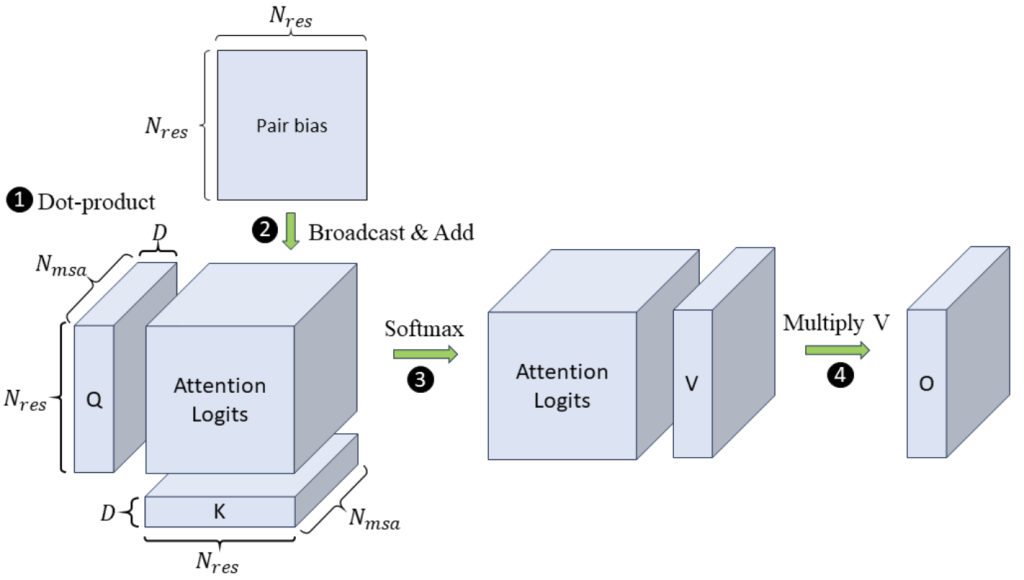
Problem Definition. The Evoformer-centric models such as OpenFold and others typically use four attention variants to process the 4D sequence tensors: MSA row-wise, MSA column-wise, and two kinds of Triangular. In particular, the input tensor is of shape , where
is the length of MSA sequences,
is the length of residue sequences,
is the number of attention heads, and
is the hidden dimension of the model. Figure 3 illustrates an example of MSA row-wise attention. The inputs consist of three projected tensors in shape
, namely
,
, and
, and a
bias matrix of residue pairs. In step 1,
and
perform dot-product between every row vector along the
dimension, deriving the attention logits in shape
as the intermediate results. For simplicity, we only depict one head in the figure. Unlike language models such as GPT-3, where
and
are considerably larger, Evoformer operates on a different scale. Specifically, MSA row-wise attention is typically designed with
heads, each having
features, while GPT-3 is configured with
heads and
features per head. However, MSA and residue sequence lengths can extend up to 5K during training and inference, respectively, making the memory explosion for intermediate results. MSA row-wise attention has the
memory footprint, and, similarly, for MSA column-wise attention, the memory footprint is
. In contrast, the memory footprint of language models is much smaller, approximately
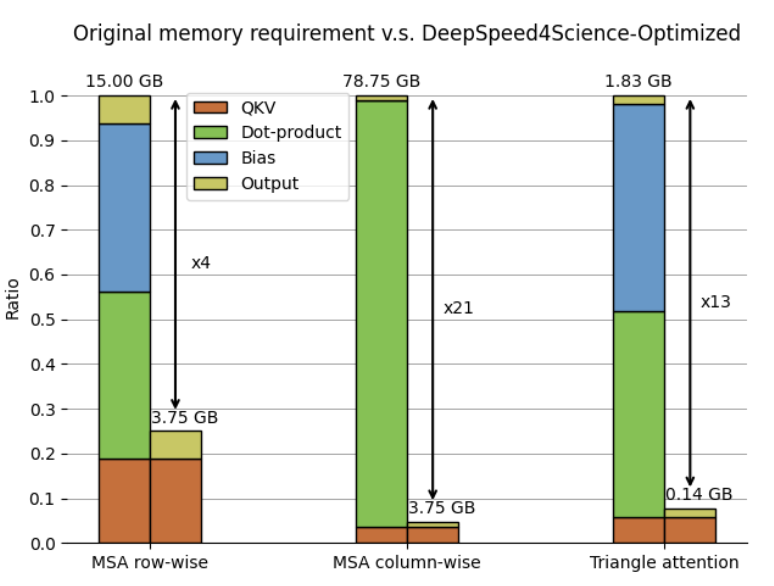
. Figure 4 shows the breakdown of memory requirements per GPU.
Existing techniques for long sequences cannot effectively address such memory explosion challenges in Evoformer’s specialized attention for structural biology. For example, MSA row-wise attention and two Triangular attention apply a bias term to the attention logits, and the bias term’s gradients are required during backward. As shown in step 2, the pair bias is derived by projecting the pair-wise representation and is used to adjust the attention logits based on the structure of residues to satisfy the spatial constraints. Take FlashAttention as an example; it cannot integrate these backward-compatible bias terms directly. Furthermore, the bias requires appropriate broadcasting to match the shape of attention logits before adding. It thus also needs to be mirrored in backward computing. Recognizing these challenges, DeepSpeed4Science addresses this memory inefficiency problem by designing customized, exact attention kernels for these attention variants in EvoformerAttention and boosting the training/inference efficiency.
Our customized highly memory-efficient DS4Sci_EvoformerAttention kernels fuses the four steps computation and calculates the attention logits in tiles. Specifically, in the forward kernel, each thread block computes a tile of in the attention logit tensor. Each thread block loads the needed tiles from
and
to perform the dot-product. The resultant tile is stored in registers and added with biases. Then, we perform softmax as step 3 and multiply V as step 4. We reduce the memory footprint by materializing only a subset of tiles in the
tensor and not saving the whole tensor for backward. We perform steps 1-3 in the backward kernel to recompute the attention logits. The backward computation is similar to that of FlashAttention. In our kernels, we tune the tile size for better performance. Large tile size leads to more efficient memory access while incurring register spilling; We tune the tile size to be
.
The bias-adding needs to be effectively broadcasted to match the bias shape with the attention logits. For example, in MSA row-wise attention, the residue pair-wise representation in shape is transformed to be the bias term in shape
, while the attention logits tensor is of shape
. To broadcast, the bias tensor will be repeated
times as the second dimension. Here, we cannot directly leverage the broadcast semantics in Pytorch because we use a fused CUDA kernel out of PyTorch. Besides, broadcasting in PyTorch requires the operation between two full tensors instead of tiles. Thus, we enabled on-the-fly broadcasting in the kernel; in particular, after calculating the attention logits after step 1. For example, a thread block loads a
tile from the pair bias. The thread block for different heads with the same position of its tile in the attention logits will load the same bias tile. The loaded tile is added to the logits tile in registers.
In backward, the gradient of the bias terms equals the gradient of attention logits. However, we need to reverse the broadcast operation. That is, the gradients along the broadcast dimension need to be accumulated. Specifically, the shape of attention logits gradients is and the bias gradient is computed similar to
in Pytorch. To reduce the memory footprint, we also fuse this operation into our kernel; otherwise, it needs the full attention logits gradient tensor. As described above, different thread blocks load the same bias tiles participants in the accumulation. Each thread block uses atomic-add operations when writing out its tile of gradients. To reduce the contention that multiple thread blocks are trying to write the same place, we schedule the thread block so that blocks executing on GPU’s multiprocessors at the same wave write to different tiles. Furthermore, the accumulation could lead to potential accuracy issues due to the round-off error of low-precision arithmetic operations, especially for bfloat16. Consequently, we convert the gradient to FP32 before adding and converting it back in another kernel if necessary. It also avoids using the slow
instruction.
How to use DS4Sci_EvoformerAttention
To use DS4Sci_EvoformerAttention in user’s own models, we need to import DS4Sci_EvoformerAttention from deepspeed.ops.deepspeed4science:
from deepspeed.ops.deepspeed4science import DS4Sci_EvoformerAttention
Take MSA row-wise attention as an example. The input tensors Q, K, V are in shape [Batch, N_msa, N_res, Head, Dim]. The pair bias is [Batch, 1, Head, N_res, N_res] and a mask to handle the padding in the batch of residue sequences with different lengths, [Batch, N_seq, 1, 1, N_res]. Simply call the kernel as
out = DS4Sci_EvoformerAttention(Q, K, V, [res_mask, pair_bias])
Detailed instructions on how to use DS4Sci_EvoformerAttention can be found at DeepSpeed4Science tutorials.
Case Study: OpenFold
Check out how OpenFold uses DS4Sci_EvoformerAttention to unblock their scientific discoveries.